¶Getting started
¶The exercise: interactive enzyme manipulation
Start up the GK-BS-v1 exercise using the graphical interface.
UnityMol will open, Biospring will start up in the background, and the
visualization will connect to the running simulation. More
instructions are provided in the video walkthrough.
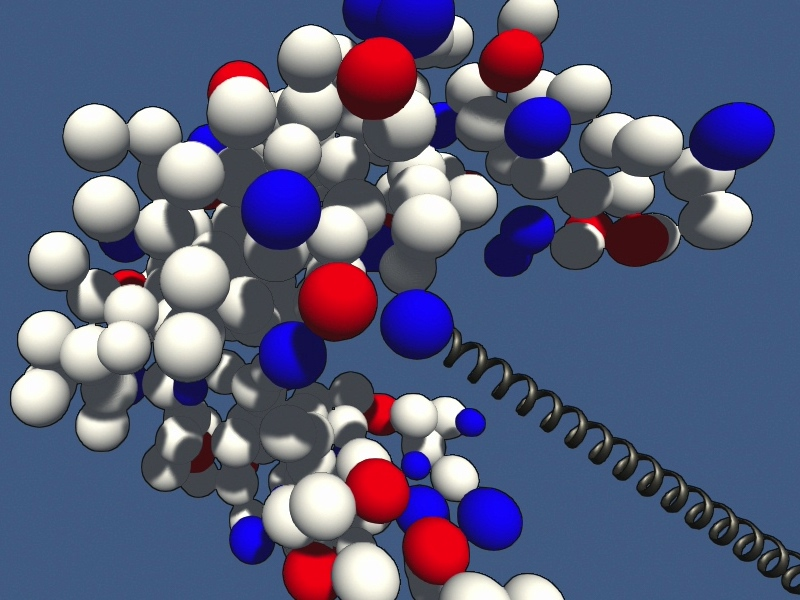
The protein is shown as multi-colored beads.
When you manipulate the simulation, be gentle, otherwise the simulation will crash and you need to start over. Common tasks may be:
- try to further open or close the binding cleft of the enzyme
- observe how applied forces propagate in this allosteric system
- identify sites that are strongly inter-connected across the structure
- compare to observations with the all-atom simulation equivalent that is provided as another demo
- trap the system in a metastable state by using the nonbonded interactions as barrier to escape
¶Minimal UnityMol tips
- on the right hand side you have a python console. You can
quit each tutorial by typing
q()plus enter. - you can modulate the applied force with the
f(value)shortcut, wherevalueis something in the range from roughly 0.1 to 6.0. You can check for the current value withfp(). - if you want to save the coordinates of the current scene, use
s(), which writessnap.pdbin the current directory (it is indicated in the console upon saving) - if you do not need the console, you can hide it with the arrow in a blue circle
- on the left hand side you have the main graphical menu. you can control the visualization, trajectory player speed and a few other settings, such as visual effects
- the menu can also be hidden with it’s attached arrow in a blue circle, when you don’t need it
¶Video Walkthrough
You’ll learn:
- Probing allosteric interactions in an enzyme
- Triggering complex long-range functional motions
- Discovering augmented elastic networks
Click like 👍 on the video if you’re excited to get started!